Note
Go to the end to download the full example code.
Training a model with ZBL corrections¶
This tutorial demonstrates how to train a model with ZBL corrections.
The training set for this example consists of a subset of the ethanol moleculs from the rMD17 dataset.
The models are trained using the following training options, respectively:
seed: 42
architecture:
name: soap_bpnn
model:
zbl: false
training:
num_epochs: 10
batch_size: 10
# training set section
training_set:
systems:
read_from: ethanol_reduced_100.xyz
length_unit: angstrom
targets:
energy:
key: energy
unit: eV # very important to run simulations
validation_set: 0.1
test_set: 0.0
seed: 42
architecture:
name: soap_bpnn
model:
zbl: true
training:
num_epochs: 10
batch_size: 10
# training set section
training_set:
systems:
read_from: ethanol_reduced_100.xyz
length_unit: angstrom
targets:
energy:
key: energy
unit: eV # very important to run simulations
validation_set: 0.1
test_set: 0.0
As you can see, they are identical, except for the zbl key in the
model section.
You can train the same models yourself with
#!/bin/bash
mtt train options_no_zbl.yaml -o model_no_zbl.pt
mtt train options_zbl.yaml -o model_zbl.pt
A detailed step-by-step introduction on how to train a model is provided in the Basic Usage tutorial.
First, we start by importing the necessary libraries, including the integration of ASE calculators for metatensor atomistic models.
import ase
import matplotlib.pyplot as plt
import numpy as np
import torch
from metatomic.torch.ase_calculator import MetatomicCalculator
Setting up the dimers¶
We set up a series of dimers with different atom pairs and distances. We will calculate the energies of these dimers using the models trained with and without ZBL corrections.
distances = np.linspace(0.5, 6.0, 200)
pairs = {}
for pair in [("H", "H"), ("H", "C"), ("C", "C"), ("C", "O"), ("O", "O"), ("H", "O")]:
structures = []
for distance in distances:
atoms = ase.Atoms(
symbols=[pair[0], pair[1]],
positions=[[0, 0, 0], [0, 0, distance]],
)
structures.append(atoms)
pairs[pair] = structures
We now load the two exported models, one with and one without ZBL corrections
calc_no_zbl = MetatomicCalculator("model_no_zbl.pt", extensions_directory="extensions/")
calc_zbl = MetatomicCalculator("model_zbl.pt", extensions_directory="extensions/")
Calculate and plot energies without ZBL¶
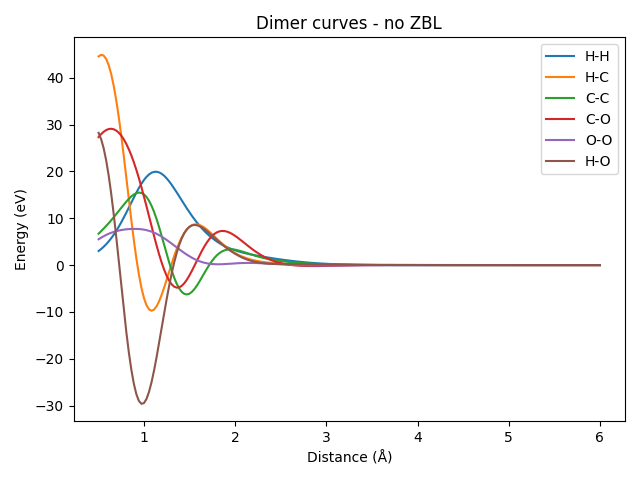
We calculate the energies of the dimer curves for each pair of atoms and plot the results, using the non-ZBL-corrected model.
for pair, structures_for_pair in pairs.items():
energies = []
for atoms in structures_for_pair:
atoms.set_calculator(calc_no_zbl)
with torch.jit.optimized_execution(False):
energies.append(atoms.get_potential_energy())
energies = np.array(energies) - energies[-1]
plt.plot(distances, energies, label=f"{pair[0]}-{pair[1]}")
plt.title("Dimer curves - no ZBL")
plt.xlabel("Distance (Å)")
plt.ylabel("Energy (eV)")
plt.legend()
plt.tight_layout()
plt.show()
/home/runner/work/metatrain/metatrain/examples/zbl/dimers.py:82: FutureWarning: Please use atoms.calc = calc
atoms.set_calculator(calc_no_zbl)
Calculate and plot energies from the ZBL-corrected model¶
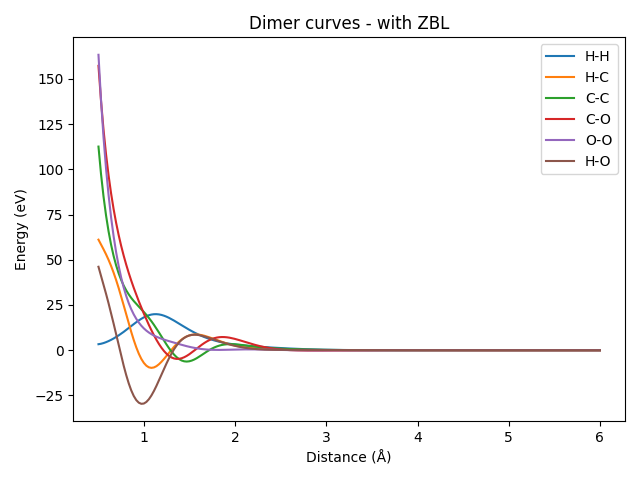
We repeat the same procedure as above, but this time with the ZBL-corrected model.
for pair, structures_for_pair in pairs.items():
energies = []
for atoms in structures_for_pair:
atoms.set_calculator(calc_zbl)
with torch.jit.optimized_execution(False):
energies.append(atoms.get_potential_energy())
energies = np.array(energies) - energies[-1]
plt.plot(distances, energies, label=f"{pair[0]}-{pair[1]}")
plt.title("Dimer curves - with ZBL")
plt.xlabel("Distance (Å)")
plt.ylabel("Energy (eV)")
plt.legend()
plt.tight_layout()
plt.show()
/home/runner/work/metatrain/metatrain/examples/zbl/dimers.py:104: FutureWarning: Please use atoms.calc = calc
atoms.set_calculator(calc_zbl)
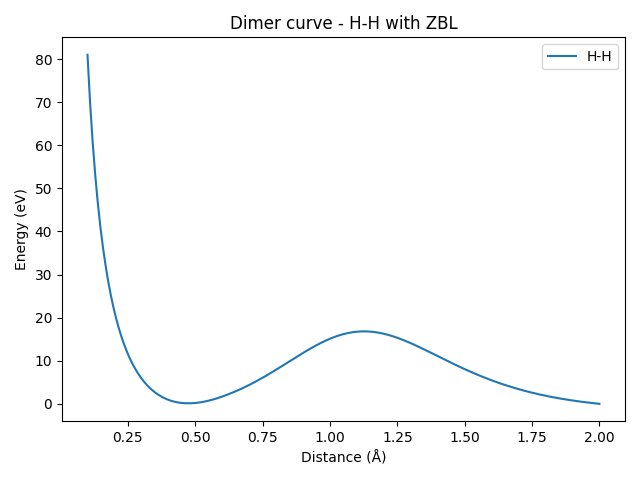
It can be seen that all the dimer curves include a strong repulsion at short distances, which is due to the ZBL contribution. Even the H-H dimer, whose ZBL correction is very weak due to the small covalent radii of hydrogen, would show a strong repulsion closer to the origin (here, we only plotted starting from a distance of 0.5 Å). Let’s zoom in on the H-H dimer to see this effect more clearly.
new_distances = np.linspace(0.1, 2.0, 200)
structures = []
for distance in new_distances:
atoms = ase.Atoms(
symbols=["H", "H"],
positions=[[0, 0, 0], [0, 0, distance]],
)
structures.append(atoms)
for atoms in structures:
atoms.set_calculator(calc_zbl)
with torch.jit.optimized_execution(False):
energies = [atoms.get_potential_energy() for atoms in structures]
energies = np.array(energies) - energies[-1]
plt.plot(new_distances, energies, label="H-H")
plt.title("Dimer curve - H-H with ZBL")
plt.xlabel("Distance (Å)")
plt.ylabel("Energy (eV)")
plt.legend()
plt.tight_layout()
plt.show()
/home/runner/work/metatrain/metatrain/examples/zbl/dimers.py:136: FutureWarning: Please use atoms.calc = calc
atoms.set_calculator(calc_zbl)
Total running time of the script: (0 minutes 38.171 seconds)