Note
Go to the end to download the full example code.
Computing LLPR uncertainties¶
This tutorial demonstrates how to use an already trained and exported model from Python. It involves the computation of the local prediction rigidity (LPR) for every atom of a single ethanol molecule, using the last-layer prediction rigidity (LLPR) approximation.
The model was trained using the following training options.
device: cpu
base_precision: 64
seed: 42
architecture:
name: soap_bpnn
training:
batch_size: 10
num_epochs: 10
learning_rate: 0.01
# Section defining the parameters for system and target data
training_set:
systems: qm9_reduced_100.xyz
targets:
energy:
key: U0
unit: hartree # very important to run simulations
validation_set: 0.1
test_set: 0.0
You can train the same model yourself with
#!/bin/bash
mtt train options.yaml
A detailed step-by-step introduction on how to train a model is provided in the Basic Usage tutorial.
import torch
Models can be loaded using the metatrain.utils.io.load_model() function. The
function requires the path to the exported model and, for some models, also the path
to the respective extensions directory. Both are produced during the training process.
%%
In metatrain, a Dataset is composed of a list of systems and a dictionary of targets. The following lines illustrate how to read systems and targets from xyz files, and how to create a Dataset object from them.
from metatrain.utils.data import Dataset, read_systems, read_targets # noqa: E402
from metatrain.utils.neighbor_lists import ( # noqa: E402
get_requested_neighbor_lists,
get_system_with_neighbor_lists,
)
qm9_systems = read_systems("qm9_reduced_100.xyz")
target_config = {
"energy": {
"quantity": "energy",
"read_from": "ethanol_reduced_100.xyz",
"reader": "ase",
"key": "energy",
"unit": "kcal/mol",
"type": "scalar",
"per_atom": False,
"num_subtargets": 1,
"forces": False,
"stress": False,
"virial": False,
},
}
targets, _ = read_targets(target_config)
from metatrain.utils.io import load_model # noqa: E402
from metatrain.utils.llpr import LLPRUncertaintyModel # noqa: E402
model = load_model("model.ckpt")
llpr_model = LLPRUncertaintyModel(model) # wrap the model in a LLPR uncertainty model
requested_neighbor_lists = get_requested_neighbor_lists(llpr_model)
qm9_systems = [
get_system_with_neighbor_lists(system, requested_neighbor_lists)
for system in qm9_systems
]
dataset = Dataset.from_dict({"system": qm9_systems, **targets})
# We also load a single ethanol molecule on which we will compute properties.
# This system is loaded without targets, as we are only interested in the LPR
# values.
ethanol_system = read_systems("ethanol_reduced_100.xyz")[0]
ethanol_system = get_system_with_neighbor_lists(
ethanol_system, requested_neighbor_lists
)
The dataset is fully compatible with torch. For example, be used to create a DataLoader object.
from metatrain.utils.data import CollateFn # noqa: E402
collate_fn = CollateFn(target_keys=list(targets.keys()))
dataloader = torch.utils.data.DataLoader(
dataset,
batch_size=10,
shuffle=False,
collate_fn=collate_fn,
)
Wrapping the model in a LLPRUncertaintyModel object allows us to compute prediction rigidity metrics, which are useful for uncertainty quantification and model introspection.
llpr_model.compute_covariance(dataloader)
llpr_model.compute_inverse_covariance(regularizer=1e-4)
# calibrate on the same dataset for simplicity. In reality, a separate
# calibration/validation dataset should be used.
llpr_model.calibrate(dataloader)
# Finally, we can save a checkpoint of the LLPR model
llpr_model.save_checkpoint("llpr_model.ckpt")
Using the LLPR model to perform uncertainty calculations requires loading it as a metatomic model. To do this, we need to export it and load the exported version.
import subprocess # noqa: E402
from metatomic.torch import load_atomistic_model # noqa: E402
# First, we run "mtt export" to export the model. This generates an exported model
# named "llpr_model.pt" and an extension directory named "llpr_extensions/".
subprocess.run(["mtt", "export", "llpr_model.ckpt", "-e", "llpr_extensions/"])
# Finally, we load the exported model using metatomic's `load_atomistic_model` function.
exported_model = load_atomistic_model(
"llpr_model.pt", extensions_directory="llpr_extensions/"
)
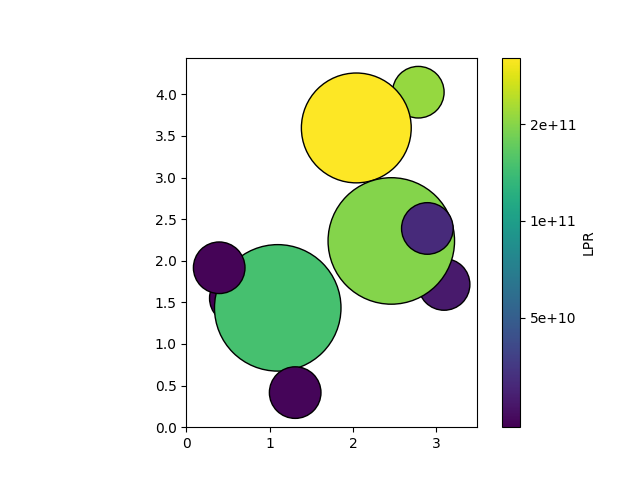
We can now use the model to compute the uncertainty for every atom in the ethanol molecule. To do so, we create a ModelEvaluationOptions object, which is used to request specific outputs from the model. In this case, we request the uncertainty in the atomic energy predictions. (Note that this “uncertainty” is in part due to the fact that the model has never been trained on per-atom energies, but only on total energies. This effect has been studied in the literature by means of the local prediction rigidity, or LPR. The LPR is the inverse of the square of the per-atom uncertainty, as defined here.)
from metatomic.torch import ModelEvaluationOptions, ModelOutput # noqa: E402
evaluation_options = ModelEvaluationOptions(
length_unit="angstrom",
outputs={
# request the uncertainty in the atomic energy predictions
"energy": ModelOutput(per_atom=True), # needed to request the uncertainties
"energy_uncertainty": ModelOutput(per_atom=True),
# `per_atom=False` would return the total uncertainty for the system,
# or (the inverse of) the TPR (total prediction rigidity)
# you also can request other outputs from the model here, for example:
# "mtt::aux::energy_last_layer_features": ModelOutput(per_atom=True),
},
selected_atoms=None,
)
outputs = exported_model([ethanol_system], evaluation_options, check_consistency=False)
one_over_lpr = outputs["energy_uncertainty"].block().values.detach().cpu().numpy() ** 2
We can now visualize the LPR values using the plot_atoms function from
ase.visualize.plot.
import ase.io # noqa: E402
import matplotlib.pyplot as plt # noqa: E402
from ase.visualize.plot import plot_atoms # noqa: E402
from matplotlib.colors import LogNorm # noqa: E402
structure = ase.io.read("ethanol_reduced_100.xyz")
norm = LogNorm(vmin=min(one_over_lpr), vmax=max(one_over_lpr))
colormap = plt.get_cmap("viridis")
colors = colormap(norm(one_over_lpr))
ax = plot_atoms(structure, colors=colors, rotation="180x,0y,0z")
custom_ticks = [2e9, 5e9, 1e10, 2e10, 5e10]
cbar = plt.colorbar(
plt.cm.ScalarMappable(norm=norm, cmap=colormap),
ax=ax,
label="LPR",
ticks=custom_ticks,
)
cbar.ax.set_yticklabels([f"{tick:.0e}" for tick in custom_ticks])
cbar.minorticks_off()
plt.show()
Total running time of the script: (0 minutes 7.824 seconds)